On the Trails of the Proteasome Fold: Structural and Functional Analysis of the Ancestral β-Subunit Protein Anbu
24-Feb-2018
JMB, Volume 430, Issue 5, Pages 628-640, https://doi.org/10.1016/j.jmb.2018.01.004
JMB, online article
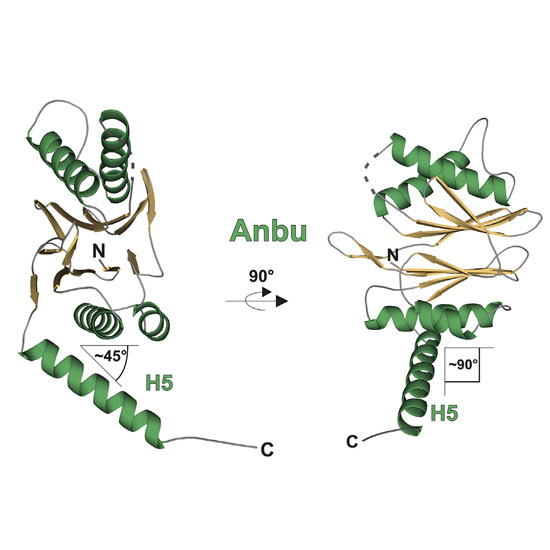
The 20S proteasome is a key player in eukaryotic and archaeal protein degradation, but its progenitor in eubacteria is unknown. Recently, the ancestral β-subunit protein (Anbu) was predicted to be the evolutionary precursor of the proteasome. We crystallized Anbu from Hyphomicrobium sp. strain MC1 in four different space groups and solved the structures by SAD-phasing and Patterson search calculation techniques. Our data reveal that Anbu adopts the classical fold of Ntn-hydrolases, but its oligomeric state differs from that of barrel-shaped proteases. In contrast to their typical architecture, the Anbu protomer is a tightly interacting dimer that can assemble into a helical superstructure. Although Anbu features a catalytic triad of Thr1Oγ, Asp17Oδ1 and Lys32Nε, it is unable to hydrolyze standard protease substrates. The lack of activity might be caused by the incapacity of Thr1NH2 to function as a Brønsted acid during substrate cleavage due to its missing activation via hydrogen bonding. Altogether, we demonstrate that the topology of the proteasomal fold is conserved in Anbu, but whether it acts as a protease still needs to be clarified.